Pseudomonas aeruginosa SN4 enhances Seedling Growth of Oryza sativa in Cadmium Contaminated Soil
Soumitra Nath1 * , Bibhas Deb2 , Indu Sharma3 and Piyush Pandey3 *
1
Bioinformatics Centre,
Gurucharan College,
Silchar,
788004
India
2
Department of Botany,
Gurucharan College,
Silchar,
788004
India
3
Department of Microbiology,
Assam University,
Silchar,
788011
India
DOI: http://dx.doi.org/10.12944/CWE.9.2.32
Copy the following to cite this article:
Nath S, Deb B, Sharma I, Pandey P. Pseudomonas aeruginosa SN4 enhances Seedling Growth of Oryza sativa in Cadmium Contaminated Soil. Curr World Environ 2014;9 (2) DOI:http://dx.doi.org/10.12944/CWE.9.2.32
Copy the following to cite this URL:
Nath S, Deb B, Sharma I, Pandey P. Pseudomonas aeruginosa SN4 enhances Seedling Growth of Oryza sativa in Cadmium Contaminated Soil. Curr World Environ 2014;9(2). Available from: http://www.cwejournal.org/?p=6367
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2014-05-24 |
|---|---|
| Accepted: | 2014-07-11 |
Pseudomonas is a genus of gram negative, nonspore forming, rod-shaped bacteria. They are commonly found in soil, water and decaying matter and including some species that are plant and animal pathogens. Pseudomonas aeruginosa is a typical example of double assessment; in a clinical environment, it is one of the most significant opportunistic pathogenic bacteria and is responsible for the majority of nosocomial infections.1 However, in the field of environmental protection, the human health concerns of this organism are not recognized, and the strains of this species are commonly used for bioremediation purposes. Pseudomonas spp. have been employed efficiently as biocontrol agents and some commercial products in the market has already shown their efficacy, nevertheless, the applications of purified siderophores, as bacteriostatic or fungistatic agents in combination with other antibacterial factors will certainly raise a great interest.Pseudomonasspp. produces numerous compounds which are responsible for disease control and also helps in the growth of plants. These inhibitory compounds are siderophores, HCN, degradative extracellular enzymes such as chitinase, protease, cellulose, β-1,3 glucanase and antibiotics such as pyrrolnitrin, pyoluteorin and phenazine.2,3,4
Materials and Methods
Collection of Soil Sample and Selective Isolation of Pseudomonas spp.
Soil samples were first collected randomly from seven different locations of Ponta Sahib, Himachal Pradesh, India. Sampling was done from garages, welding shops, petrol pumps and other polluted sites. Samples were collected in sterilized polythene bags after proper labeling and immediately bought to the laboratory.Soil plating was done within 72 hrs. of sample collection. Selective isolation of Pseudomonas spp. was done by spreading the samples on Pseudomonas Isolation Agar (PIA)media and incubatingat 37ºC for 24 hrs. Pure cultures were finally obtained by repeated sub-culturing followed by cadmium tolerance tests.
Characterization of the bacterial isolates
Pure culture of Pseudomonas spp. that were tolerant to cadmium were identified up to species level by their morphological and biochemical characterization5,6 and finally confirmed by 16S rDNA sequencing.
Identification by 16S rDNA sequencing
Pure cultures were grown until log phase and genomic DNA was extracted from bacterial isolates.7 The amplification of 16S rRNA gene was done by using forward primer and reverse primer. The ~1.4 kb-PCR products of 16S rRNA genes were used for DNA sequencing. After sequencing, the sequence was analysed by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and Ribosomal Database Project (http://rdp.cme.msu.edu/) for finding the closest homologous sequence. The first ten homologous sequences were selected based on their maximum identity score. The sequences were than aligned and a distance matrix was constructed followed by the construction of a phylogenetic tree byNeighbour-Joining method.
Screening for Cadmiumtolerance
All the isolates were checked for metal tolerance. Minimum Inhibitory Concentration (MIC) was determined against cadmium (CdCl2) by gradually increasing the concentration of the cadmium on Nutrient Agar (NA) plates until the strains failed to give colonies on the plate. The initial concentration was 50μg/ml and the cultures grown on last concentration was transferred to the higher concentration by streaking on the plate. MIC was recorded when the isolates failed to grow on plates. Minimum inhibitory concentration for copper, lead and zinc were also determined.
Antibiotic Sensitivity and Resistance Pattern of the Recovered Isolates
The isolates were tested for antibiotic sensitivity according to Kirby-Bauer disc diffusion method8 and the antibiotics discs were procured from ‘HIMEDIA’. The antibiotics that were used in the present study are Amikacin, Amoxycillin, Ampicillin, Cefalexin, Cefixime, Ceftriaxone, Chloramphenicol, Gentamicin, Kanamycin, Methicillin, Ofloxacin and Tetracycline. The diameter of the inhibition zones was measured to the nearest mm and the isolates were classified as resistant (R), intermediate (I) and susceptible (S) following the standard antibiotic disk sensitivity testing method.
Pot Experiment Preparation of Bacterial Inoculum
Pseudomonas aeruginosa SN4, which showed a greater degree of tolerance for cadmium, was taken under consideration for the preparation of bacterial inoculum. The isolate was inoculated in nutrient broth and kept in shaker incubator at 120 rpm at 28 ± 2°C for 48 hours. After incubation period, 5 ml of broth was added to 45 ml distilled water for the formulation of biofertilizer and to carry out the pot experiment.
Collection and Pre-Sowing Treatment of Oryza Sativa Seedlings
Seeds of Oryza sativawere collected from KrishiVikas Kendra, Masimpur, Assam. The size and weight of seeds were homogenous. Clean seeds were dipped in water; floating seeds were discarded, while seeds settled on bottom of container were selected. Seeds were surface-sterilized with 95% alcohol for 30 seconds, followed by 0.1% (w/v) HgCl2 for 1–2 min and then washed with sterile distilled water for 5–6 times.9 The seeds were then put in a sterile petridish containing Hogland Solution and kept overnight.
Pot Trial Studies
The earthen pots (24 cm X 12 cm X 12 cm) were filled with sterilized sandy loam soil. Seeds were sown on all the pots to study the role of P. aeruginosa SN4on shoot growth of Oryza sativa sown in cadmium incorporated soil (at 20 mg/kg and 50 mg/kg of soil). Pot experiment was performed by adding the biofertilizer formulation to cadmiumcontaminated soil. Seedling germinationand its growth pattern was recorded everyday for three weeks.
Statistical Analysis
After performing the pot experiment, SPSS 16.0 was used to analyze the statistical data. Descriptive statistics calculates the means of all replicates with standard error and deviations. Multiple comparison tests were performed to evaluate the effectiveness of each bacterial isolates. When analysis of variance (ANOVA) showed significant effects, Tukey’s-b test (assuming equal variances) and Games-Howell test (assuming unequal variances) was done to make comparison between groups at P<0.05 and P<0.01.
Results Characterization of Selected Isolates
A total of thirty two Pseudomonas spp. were isolated fromPonta Sahib, Himachal Pradesh, India. Total viable counts ranges from 32 X 104 (CFU/g) to 43 X 104 (CFU/g). All the isolates were gram negative rod. Most of the isolates were found to be indole negative, MR negative, VP negative and urease negative. Oxidase tests for Pseudomonas isolates were positive indicating them to be aerobic strains.Positive result for citrate test was observed, infers the ability of these organisms to utilize citrate as the sole source of carbon and energy. Isolates were also able to produce an enzyme “nitrate reductase” resulting in the reduction of nitrate (NO3). The isolate which showed highest tolerance for cadmium and also showed significant result in pot experiment was selected for 16S rDNA sequencing. A neighbour-joining tree was generated using the sequence from Ps-11 (1393 bp) and representative sequences from databases. It has been observed that the strain code Ps-11 had maximum sequence similarity with the species of Pseudomonas aeruginosa and occupied the same phylogenetic branch (figure 1). The sequence has been identified as Pseudomonas aeruginosa SN4 and submitted to NCBI-GenBank. The accession number thus obtained is KF44770.
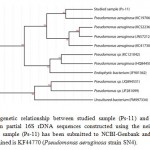 |
Figure 1: Phylogenetic relationship between studied sample (Ps-11) and representative species based on partial 16S rDNA sequences constructed using the neighbour-joining method. Studied sample (Ps-11) has been submitted to NCBI-Genbank and the accession number thus obtained is KF44770 (Pseudomonas aeruginosa strain SN4). Click here to View Figure |
Minimum Inhibitory Concentration (MIC) of the recovered isolates
All the bacterial isolates exhibited resistance to cadmium and the minimum inhibitory concentration (MIC) rangesfrom 300μg/ml to 1800μg/ml. There is a great variation in the tolerance capacity of isolated bacterial strains against cadmium chloride. Around 53% of the tested isolates were tolerant at 1000μg/ml CdCl2 concentration. Pseudomonas aeruginosa SN4 (Isolate code: Ps-11)exhibited high resistance with MIC for cadmium as 1800μg/ml. Multi-metal tolerance test showed that the MIC value for P. aeruginosa SN4 for lead was 170μg/ml, copper was 60μg/ml and zinc was 1800μg/ml (table 1).
Table 1: Minimum inhibitory concentration of all the isolated strains
|
Isolate code |
Minimum Inhibitory Concentration |
|||
|
Cadmium |
Lead |
Copper |
Zinc |
|
|
Ps-1 |
1400 µg/ml | 150 µg/ml | 50 µg/ml | 1800 µg/ml |
|
Ps-2 |
1100 µg/ml | 80 µg/ml | 60 µg/ml | 1500 µg/ml |
|
Ps-3 |
800 µg/ml | 100 µg/ml | 40 µg/ml | 1600 µg/ml |
|
Ps-4 |
700 µg/ml | 110 µg/ml | 30 µg/ml | 1100 µg/ml |
|
Ps-5 |
1000 µg/ml | 150 µg/ml | 40 µg/ml | 1600 µg/ml |
|
Ps-6 |
1200 µg/ml | 160 µg/ml | 50 µg/ml | 1700 µg/ml |
|
Ps-7 |
700 µg/ml | 120 µg/ml | 50 µg/ml | 1200 µg/ml |
|
Ps-8 |
400 µg/ml | 100 µg/ml | 30 µg/ml | 1000 µg/ml |
|
Ps-9 |
1100 µg/ml | 140 µg/ml | 20 µg/ml | 1200 µg/ml |
|
Ps-10 |
1000 µg/ml | 150 µg/ml | 50 µg/ml | 1300 µg/ml |
|
Ps-11 |
1800 µg/ml | 170 µg/ml | 60 µg/ml | 1800 µg/ml |
|
Ps-12 |
600 µg/ml | 100 µg/ml | 60 µg/ml | 1000 µg/ml |
|
Ps-13 |
1700 µg/ml | 160 µg/ml | 60 µg/ml | 1600 µg/ml |
|
Ps-17 |
1400 µg/ml | 170 µg/ml | 60 µg/ml | 1800 µg/ml |
|
Ps-15 |
300 µg/ml | 100 µg/ml | 30 µg/ml | 1000 µg/ml |
|
Ps-16 |
500 µg/ml | 120 µg/ml | 20 µg/ml | 900 µg/ml |
|
Ps-17 |
1200 µg/ml | 170 µg/ml | 60 µg/ml | 1300 µg/ml |
|
Ps-18 |
800 µg/ml | 140 µg/ml | 50 µg/ml | 1000 µg/ml |
|
Ps-19 |
800 µg/ml | 150 µg/ml | 40 µg/ml | 1300 µg/ml |
|
Ps-20 |
600 µg/ml | 120 µg/ml | 30 µg/ml | 900 µg/ml |
|
Ps-21 |
1200 µg/ml | 150 µg/ml | 60 µg/ml | 1400 µg/ml |
|
Ps-22 |
600 µg/ml | 120 µg/ml | 50 µg/ml | 1300 µg/ml |
|
Ps-23 |
400 µg/ml | 120 µg/ml | 40 µg/ml | 1300 µg/ml |
|
Ps-24 |
400 µg/ml | 120 µg/ml | 40 µg/ml | 1000 µg/ml |
|
Ps-25 |
1500 µg/ml | 130 µg/ml | 40 µg/ml | 1200 µg/ml |
|
Ps-26 |
1200 µg/ml | 120 µg/ml | 50 µg/ml | 900 µg/ml |
|
Ps-27 |
1800 µg/ml | 170 µg/ml | 60 µg/ml | 1800 µg/ml |
|
Ps-28 |
1000 µg/ml | 130 µg/ml | 40 µg/ml | 1500 µg/ml |
|
Ps-29 |
1200 µg/ml | 120 µg/ml | 30 µg/ml | 1200 µg/ml |
|
Ps-30 |
600 µg/ml | 120 µg/ml | 30 µg/ml | 900 µg/ml |
|
Ps-31 |
300 µg/ml | 100 µg/ml | 30 µg/ml | 1000 µg/ml |
|
Ps-32 |
1000 µg/ml | 130 µg/ml | 40 µg/ml | 1500 µg/ml |
Antibiotic sensitivity and resistance pattern of Pseudomonas aeruginosa SN4
Most of the Pseudomonas spp. isolated in the present studyexhibites high resistance pattern towards a group of antibiotics. It has been observed that P. aeruginosa SN4 were resistant to amoxycillin, ampicillin, cefalexin, cefixime, kanamycin, methicillin and tetracycline.This fact was also established by other researchers that multiple metal resistance bacterial isolates exhibits high resistance towards a group of antibiotics.10
Effect of Pseudomonas aeruginosa SN4 on growth of Oryza sativa inoculated in cadmium incorporated soil
Inoculation with P. aeruginosa SN4significantly increases seedling germination and growth of Oryza sativa when compared with control sets. After 20 days of seedling germination, multiple comparison result reveals that P. aeruginosa SN4 attains 12% increased seedling growth as compared to uninoculated control pots at 50mg/kg concentrations of cadmium in soil (table 2). However, at 20mg/kg cadmium in soil, a slight increased germination (mean difference = 0.96 ± 0.63 cm)was observed which was statistically non-significant. The result demonstrates the maximum bio-absorption of cadmiumby P. aeruginosa SN4at elevated levels of cadmium in soil.
Table 2: Seedling growth of Oryza sativa inoculated with cadmium resistant P. aeruginosa SN4 in cadmium incorporated soil
| Experimental setup |
Shoot length (in cm) |
|
|
Cd at 20mg/kg soil |
Cd at 50mg/kg soil |
|
| Control (without Cd and P. aeruginosa SN4) | 31.66 ± 0.42 | 31.66 ± 0.42 |
| Uninoculated control (with Cd only) | 27.62 ± 0.35 | 25.18 ± 0.26 |
| P. aeruginosa SN4 + Cd | 28.58 ± 0.98ns | 28.18 ± 0.49* |
Values are mean ± standard deviation of five replicates; ns= non significant; *= significant at P<0.01; compared with uninoculated control.
Discussion
Increased concern over the impact of cadmium on our environment, especially the vegetation has resulted in the increased interest in bioremeditation strategies. Microscopic evidence showed that isolated bacteria weregram- negative, rod-shaped. Further, biochemical tests and 16S r DNA confirms the isolates as Pseudomonas aeruginosa. A decrease in growth (cfu/g) of bacterial colonies was observed on increasing the cadmium concentration on culture plates at any given time interval compared to the control without metal amendment. The lower values of microbial load at higher metal concentrations showed correlation with the study of Anyanwuet al.11. Present study demonstrated the cadmium resistance pattern of Pseudomonas isolates. Based on the MIC values and antibiogram pattern of the isolated strains and as studied by Bruins et al.,12, Pseudomonas sp. shows resistance to a variety of toxic substances, heavy metal and antibiotics. Pseudomonas aeruginosaSN4 can tolerate upto 1800μg/ml cadmium and also showed multi-metal tolerant activity. The toxic levels of heavy metals effect structural and permeability properties of inner membranes and organelles, that results in inhibition of enzymatic activities, nutrient imbalances, decreases in rates of photosynthesis and transpiration13,14, stimulate formation of free radicals and reactive oxygen species resulting in oxidative stress15, suppress seed germination and seedling growth, reproductive development, seed yield and seed quality16 and induce deleterious anatomical and ultra-structural changes in crop plants17,18.In the present study, application of Pseudomonas aeruginosaSN4 showed a remarkable increase in seedling growth and germination when sown in cadmium incorporated soil (at both 20 and 50mg/kg). After 20 days of seedling inoculation in Cd incorporated soil and application P. aeruginosa SN4, it has been observed that rice plant attains a significantseedling growth as compared to uninoculated control pots. Several studies have evidenced the fact that cadmium-resistant and plant growth-promoting bacteria can protect plants from the toxic effects of metals19,20,21. Overall experiment demonstrated that P. aeruginosaSN4 (Genbank Acc. No: KF44770) could increase the growth of Oryza sativa in cadmium contaminated crop field (figure 2), thus dedicating sites which are set aside for long term agricultural purpose. Further research is still required to expand the knowledge of the P. aeruginosa SN4 before being used for commercial purpose.
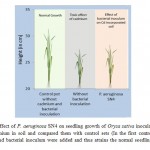 |
Figure 2: Effect of P. aeruginosa SN4 on seedling growth of Oryza sativa inoculated in 50 mg/kg cadmium in soil and compared them with control sets (In the first control pot, no cadmium and bacterial inoculum were added and thus attains the normal seedling growth. However, in the second pot, cadmium was added at concentration 50 mg/kg in soil; demonstrates the toxic effect of cadmium in seedling germination and growth. In the final test pot, addition of Pseudomonas aeruginosa strain SN4 results in attaining a significant seedling growth as illustrated in table 2). Click here to View Figure |
Conclusion
Present study isolated 32 cadmium tolerant bacteria fromcontaminated sites of Ponta Sahib, Himachal Pradesh, India. Most of the isolates exhibited multi-metal tolerance and were resistant to a group of antibiotics.P. aeruginosa SN4 showed the highest tolerance for cadmium and was taken under consideration for pot experimental studies. Comparedwith control treatments, inoculation with P. aeruginosa SN4influences the seedlinggermination at 50mg/kg cadmium concentration in soil. It has been observed that, microbial inoculation altered the bioavailability of metals in soil, resulting in 12% increased seedling growth after 20 days of bacterial inoculation.It is evident from the present study that the application of HMRB specifically adapted to high concentrations of heavy metals will increase the ability to remediate heavy metal contaminated soils. Further research is still required to evaluate the bio-absorption potential of P. aeruginosa SN4 by field trials and the underlying mechanism of action.
Acknowledgements
The authors wish to extend their grateful thanks to Department of Biotechnology, Govt. of India, New Delhi for the establishment of Institutional Level Biotech Hub and Bioinformatics Centre in Gurucharan College, Silchar, India. Authors also extend their grateful thanks to Himachal Institute of Life Sciences, Ponta Sahib, H.P, India for providing help in the initial phase of the present study.
References
- Azadeh B.F. and Meon S., Am. J. Applied Sci., 6: 1915-1919,(2009)
- Dowling D.N. and O’Gara F., Treads Biotechnol.,12: 133-141 (1994).
- Haas D. and Défago G., Nat Rev Microbiol., 3: 307-319 (2005).
- Deshwal V.K., Devi M.S., Bhajanka N., Mistri J., Bose A. and Saini N., Global J ApplAgr Res., 1: 49-55 (2011).
- Cappuccino J.G. and Sherman N.,Microbiology: A Laboratory Manual, 7th Ed, pp 161-204. Pearson, India (2005).
- Harley J.P., Laboratory exercises in Microbiology, 6th ed. McGraw Hill, New York, NY (2005).
- Sambrook J. and Russell D.W., Molecular Cloning: A Laboratory Manual, 3rd Ed. Cold Spring Harbor Laboratory Press(2001).
- Bauer A.W., Kirby W.M.M., Sherris J.C. and Turk M., Am.Jl of Clinical Path., 45: 493–496 (1966).
- Singh N., Pandey P.,Dubey R. C. and Maheshwari D. K., World J MicrobiolBiotechnol, 24: 1669–1679 (2008).
- Vajiheh K., Naser B. and Giti E., African J. Biotechnol., 2:379-383 (2003).
- Anyanwu C.U., Nwankwo S.C. and Moneke A.N., Int. J of Basic& App Sci.,11:109-115 (2011).
- Bruins M.R., Kapil S. and Oehme F.W.,Ecotox. Environ. Safe.,54: 241-248 (2003).
- Green C., Vhaney R. and Bouwkamp J.,J. Plant Nutr.,26: 417-430 (2003).
- Azevado H., Pinto C.G.G., Farnandes J., Loureiro S. and Santos C.,J. Plant Nutr.,28: 2211-2220 (2005).
- Sandalio L.M., Dalurzo H.C., Gomez M., Romero-Puertas M.C. and del Rio L.A.,J. Expt. Bot., 52: 2115-2126 (2005).
- Beri A. and Setia R.C.,J Indian Bot. Soc., 74: 293-297 (1995).
- Liu D. and Kottke I., J. Biosci., 29: 329-335 (2004).
- Maruthi Sridhar B.B., Diehl S.V., Han F.X., Monts D.L. and Su Y., Environ. Expt. Bot., 54: 131-141 (2005).
- Burd G. I., Dixon D. G., and Glick B. R., Can. J. Microbiol., 46: 237-245 (2000).
- Dell’Amico E., Cavalca L. and Andreoni V.,FEMS Microbiol. Ecol., 52: 153-162 (2005).
- Jiang C.Y., Sheng X.F., Qian M. and Wang Q.Y., Chemosphere.,72: 157–164 (2008).






