Studies of environmental water samples for different parameters for isolation of Legionella spp.
Shubhra Shukla1 * , Sanjeev Kumar Shukla1 , Jose Mathew1 and Deepak Sharma1
DOI: http://dx.doi.org/10.12944/CWE.4.2.08
Legionella pneumophila has frequently been isolated from patients with Legionnaires disease and in several instances also from epidemic-related environmental samples. A selective medium was developed and used successfully to isolate Legionella pneumophila and Legionella-like organisms from environmental specimens animal inoculation methods. Legionella pneumophila is a Gram-negative, facultative, intracellular bacterium that is the causative agent of Legionnaires' disease. This medium consists of charcoalyeast extract agar to which have been added cephalothin (4µg/mL), colistin (16µg/mL), vancomycin (0.5µg/mL), and cycloheximide (80µg/mL). Pretreating of the environmental water samples with an acid buffer (pH 2.2), followed by plating on the selective medium, improved the rate of recovery of both Legionella and Legionella-like organisms relative to that with direct plating on selective media.
Copy the following to cite this article:
Shukla S, Shukla S.K, Mathew J, Sharma D. Studies of environmental water samples for different parameters for isolation of Legionella spp. Curr World Environ 2009;4(2):313-320 DOI:http://dx.doi.org/10.12944/CWE.4.2.08
Copy the following to cite this URL:
Shukla S, Shukla S.K, Mathew J, Sharma D. Studies of environmental water samples for different parameters for isolation of Legionella spp. Curr World Environ 2009;4(2):313-320. Available from: http://cwejournal.org?p=196/
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2009-08-02 |
|---|---|
| Accepted: | 2009-09-30 |
Introduction
These include air conditioning cooling towers or evaporative condensers1-4. It has also been isolated from surface water from a stream, from mud from a stream bottom5, and from lake waters not associated with epidemics. In its natural environment, the bacterium resides within freshwater amoebas such as Acanthamoeba and Hartmenella, but the organism can also infect humans upon the inhalation of contaminated aerosols. Invasion and replication in alveolar macrophages can lead to pneumonia or "Legionnaires' disease" in individuals who are immunocompromised or have some underlying conditions that reduce the efficiency of their lung defense mechanisms6.
These data suggest that Legionella secretes type IV effectors into the host cell cytosol that function to target specific host trafficking pathways in order to remodel the nascent vacuole7,8. L. pneumophila has been detected only sporadically by use of the cultivation method in untreated groundwater (GW) and in treated water at temperatures below 20°C 9-11. Primary macrophages derived from bone marrow of the permissive A/J mouse, the interaction with the lysosomal pathway is only delayed, as the RV eventually matures into an acidified vacuole in a process that resembles autophagy12.
In addition to L. pneumophila, the agent of Legionnaires disease, there are now four other proposed species of Legionella13: Legionella micdadei (pittsburgensis), Legionella dumoffii, Legionella bozemanii, and Legionella gormanii. All of these species but L. gormanii have been isolated as causative agents for illnesses involving pneumonia14,15.
However, it is not clear if these or other organisms represent potential pathogens, because representatives of L. pneumophila serogroups and genotypes also show distinct differences in infectivity. The semiselective buffered charcoal yeast extract (BCYE) agar medium supplemented with antibiotics was used to detect culturable Legionella16, 17.
Strains of L. micdadei were isolated from blood from patients. L. micdadei has also been isolated from patients and shown to be the causative agent of pneumonia18, 19. The organism has also been isolated from environmental sourcesspecifically, nebulizers from respiratory therapy equipment20. L. pneumophila is responsible for more than 90% of the reported cases of legionellosis21. Currently, about 50 Legionella species have been defined, nearly half of which have been associated with cases of disease (e.g., L. pneumophila, L. micdadei, L. bozemanii, L. longbeachae, and L. dumoffii)22.
The ability of Legionella spp. to multiply in biofilms in low-pH environments has recently been demonstrated, and those authors also concluded that water temperature affected species composition23. L. gormanii was isolated from soil collected from a creek bank during a Legionnaires disease outbreak investigation. The relationship of this organism to Legionnaires disease or other human diseases is not known at this time, although there is evidence that it can cause human as well as animal infections24. These isolates, referred to as Legionella-like organisms (LLO).
The procedure currently used to isolate L. pneumophila and other Legionella spp. The organism can be isolated either from the guinea pig tissues or the egg yolk sacs on charcoal-yeast extract (CYE) agar plating medium25. In addition to being tedious, this method is costly in terms of laboratory animals, eggs, and time. The procedure may take several weeks and does not allow for rapid processing of specimens, which might be of critical importance during an epidemic investigation.
This report describes a selective isolation procedure that has been used to isolate Legionella directly from environmental samples without animal or egg inoculation. This procedure utilizes a selective medium consisting of CYE agar with four antibiotics added: cephalothin, colistin, vancomycin, and cycloheximide. L. pneumophila is relatively resistant to low pH for a short period of time26. Our results show that the rate of recovery of Legionella from environmental water samples is improved if samples are treated with acid and then plated on the selective agar medium.
Materials and Methods
Bacterial strains
A stock culture of L. pneumophila that had been passed on artificial media several times was used for plate counts and antibiotic minimum inhibitory concentration determinations. This strain is referred to as the "stock strain" in this report. A 48-h CYE agar culture of the stock strain was suspended in yeast extract broth with 40% glycerol, divided into small aliquots, and frozen at -70°C. A vial was thawed and streaked on CYE agar for use each week.
Another culture of L. pneumophila, referred to in this report as the "tissue strain," had never been passed on artificial media. The tissue strain was stored as macerated spleen tissue from a guinea pig infected with the disease. The infected spleen, after having been macerated, was divided into small aliquots and frozen at -70°C. For each experiment, a vial was thawed and diluted 1:5,000 in phosphate buffered saline before being plated on the media to be tested.
Environmental water samples
The environmental samples used in this study were from various parts of the Uttar Pradesh and were specimens from which Legionella or LLO had already been isolated with the guinea pig/egg inoculation method. Some of the specimens were collected during epidemic investigations of Legionnaires disease outbreaks but may not havebeen related to the outbreaks. The samples were stored at 4°C for 2 to 18 months before being used in the study.
Media
The CYE agar used in this study contained 1.0% yeast extract (Hi-Media ), 1.7% agar (Biogene), 0.15% activated charcoal, 0.04% Lcysteine hydrochloride, and 0.025% ferric pyrophosphate (soluble). The pH was adjusted to 6.9 to 7.0 after autoclaving and the addition of Lcysteine and ferric pyrophosphate. Selective media were prepared by adding solutions of antibiotics to autoclaved CYE agar held in a water bath at 50°C until immediately before the plates were poured.
Yeast extract broth consisted of a 1.0% yeast extract (Hi-Media) solution to which 0.04% Lcysteine hydrochloride and 0.025% ferric pyrophosphate were added after autoclaving. The pH of the broth was adjusted to 6.9 to 7.0. FG agar (Mueller-Hinton, with 0.04% L-cysteine hydrochloride and 0.025% ferric pyrophosphate, pH 6.9 to 7.0 was used to detect the production of brown pigment by isolates presumed to be Legionella. Another medium, yeast extract-tyrosine agar, consisting of 1.0% yeast extract (Hi Media), 1.7% agar (Biogene), and 0.04% L-tyrosine, was also used for detection of brown pigment production since many organisms that were difficult to grow on FG agar grew satisfactorily on yeast extracttyrosine agar.
Direct plating of environmental samples
Two 10-fold dilutions of each environmental specimen were prepared in phosphate-buffered saline (pH 7.2 to 7.3). A 0.1-ml quantity of each dilution and of the undiluted specimen was spread onto the surface of CYE and selective agar plates in duplicate. The plates were incubated at 35°C in 2.5% C02. The plates were read after 5, 6, 7, and 10 days of incubation for colonies with morphology similar to that of Legionella (white, glistening, convex, circular, entire, and from 1 to 2 mm in diameter with a "ground glass" appearance under magnification). At the time of each reading, all colonies thought to be Legionella were subcultured to fresh CYE agar. A total plate count of all colonies was recorded at 10 days.
Acid buffer treatment of environmental samples
The HCl-KCl buffer used for pretreating water samples was prepared by mixing 3.9 ml of 0.2 M HCl with 25 ml of 0.2 M KCl to yield a buffer solution with a pH of approximately 2.2. Two procedures designated "A" and "B" for acid treatment were used. In procedure A, 10 ml of the water sample was centrifuged at 4,000 rpm for 10 min in aerosol-free centrifuge containers. The supernatant was poured off, and the sediment was resuspended in 1 ml of the original water sample. This suspension was diluted 1:10 with the HCl-KCl buffer. At intervals ranging from 5 to 60 min, 0.1-ml quantities were removed from the acid suspension and were plated on duplicate agar plates of CYE and selective media. After each such interval, a sample was diluted 1:10 with phosphate-buffered saline and then plated in 0.1-ml quantities on duplicate CYE and selective media plates.
Procedure B did not involve centrifugation but involved adding 0.5 ml of the water sample to 4.5 ml of HCl-KCl buffer. At intervals of 5 to 60 min, 0.1-ml quantities of undiluted and diluted (1:10 in phosphate buffered saline) acid-treated water sample were plated onto CYE and selective media. This procedure resulted in plating a 10 and a 10-2 dilution of the water sample, which was 10-fold more dilute when plated than samples prepared according to procedure A.
The plates inoculated with the acid-treated suspensions were incubated, read, and counted as described above for direct plating of water samples.
Screening suspect Legionella isolates
Each colony thought to be Legionella was streaked for isolation to a CYE agar plate (containing no antibiotics) and incubated for 2 to 5 days. From each such plate, the isolate was streaked onto yeast extract-tyrosine agar (or FG) and 5% sheep blood agar. If growth did not occur on blood agar after 5 days of incubation (Legionella characteristically fails to grow on blood agar) or if the isolate produced browning of the yeast extract-tyrosine agar or FG agar, the isolate was examined by the fluorescentantibody (FA) staining method with conjugated antisera to each of the five species ofLegionella. Isolates that were positive by FA in the polyvalent L. pneumophila conjugate were then further examined by FA with type-specific conjugates to L. pneumophila serogroups 1 to 6. If the FA results indicated a Legionella species, the isolate was examined for the cellular fatty acids characteristic of the Legionella species to confirm the identification, If the fatty acid profile demonstrated a pattern ofa high percentage of branched-chain cellular fatty acids, and if the isolate did not grow on blood agar or produce browning on FG or yeast extract tyrosine agar, was gram negative, and was otherwise similar to Legionella, the isolate was considered to be an LLO27, 28.
Selective media experiments
To determine their suitability for inclusion in a selective medium, colistin and cephalothin were added separately to CYE agar in several different levels. Plate counts of the L. pneumophila stock strain were performed on these selective media andon CYE agar with no antibiotics. In addition, 10 environmental samples were direct plated on these media as described above. Subsequently, cephalothin at a level of4μg/ml was combined in CYE agar with different levels of colistin (2, 4, 8, 16, and 32μg/ml) to determine the optimum concentration. The L. pneumophila stock strain and two environmental specimens were plated on these five selective media with cephalothin and and 16μgof colistin per ml was selected as optimal, and tothis medium were added two concentrations of vancomycin (3 and 0.5μg/ml). On these two selective media, and also on CYE medium, plate counts of the stock strain of L. pneumophila were done. Using cephalothin-colistinvancomycin (CCV) agar and CYE agar, five environmental water samples were plated by using the method described.
Edelstein-Finegold (E-F) medium (CYE agar with 40 U of polymyxin B and 0.5μg of vancomycin per ml) was compared with CCV medium (CYE agar with 4μg of cephalothin, 16μg of colistin, and 0.5μg of vancomycin per ml) for isolation of Legionella and suppression of non- Legionella organisms. Ten environmental water samples from which Legionella had been isolated by guinea pig/egg inoculation were used. Samples were diluted and plated as described above on three media: E-F medium, CCV medium, and CYE medium without antibiotics. These plates were read and picked for Legionella colonies, and total plate counts were determined to assess the inhibition of total flora.
In a final attempt to improve the selectivity of CCV medium, we added cycloheximide (an antifungal agent) because the prolonged incubation time (10 days) necessary to recover Legionella can sometimes result in overgrowth by fungi. A comparison of plate counts of the stock and tissue strains of L. pneumophila on CCV medium containing various concentrations of cycloheximide resulted in the selection of 80μg of cycloheximide per ml as the optimum level. Thirteen environmental water samples that had already been shown to bepositive for Legionella and LLO were plated on CYE agar with, per ml, 4μg of cephalothin, 16μg of colistin, 0.5μg of vancomycin, and 80μg of cycloheximide (CCVC medium) and on CYE agar without antibiotics.
Acid treatment experiments
To evaluate the effects of low pH on survival of Legionella over a period of time, we exposed the stock strain of L. pneumophila to the HCl-KCl buffer (pH 2.0) for periods of 5, 15, 30, and 60 min. Plate counts of the acid suspension of the stock strain were performed after each time interval.
Five water samples already shown to be positive for L. pneumophila by the guinea pig/egg inoculation method were subjected to the HCl-KCl buffer treatment for 30 min according to procedure A, and attempts were made to recover Legionella. Two naturally contaminated environmental water samples, LS- 4 and MI-56, were retested using 0-, 5-, 15-, and 30- min exposure times to HCl-KCl buffer to determine the most appropriate exposure interval for acid buffer treatment.In the last experiment the acid buffer treatment (procedures A and B), with an exposure time of 5 min and with samples directly plated onto CYE medium and CCVC selective medium, were compared for efficacy in isolating Legionella from 11 environmental water samples already shown to be positive for Legionella by the guinea pig/eg inoculation method.
Results
The growth of the L. pneumophila stock strain was inhibited on CYE medium containing 8 or 16μg of cephalothin per ml, although the minimum inhibitory concentration of cephalothin for L. pneumophila has been reported to be 16μg/ml21, 22. Although the minimum inhibitory concentration of colistin was reported to be 3.6μg/ml, colistin did not inhibit growth of L. pneumophila (stock and tissue strain) even at a level of 7.2μg/ml in our study. If an antibiotic is to be an effective agent in selective media it must inhibit bacteria other than Legionella, however, colistin at 7.2μg/ml did not sufficiently reduce the total plate counts of 10 environmental specimens as compared to the total plate counts on CYE without colistin. The average total plate count with no antibiotic was 7.7 X 105, and that with 7.2μg of colistin per ml was 2.5 X 105.
Using cephalothin-colistin medium (CYE agar with 4μg of cephalothin and 16μg of colistin per ml), we isolated L. pneumophila group 1 from 1 to 2 environmental samples, but the average amount of suppression of competing non-Leginella flora was less than 1 log. CCV medium (CYE agar with, per ml, 4μg of cephalothin, 16μg of colistin, and 0.5μg of vancomycin) was more inhibitory for grampositive spreading colonies than cephalothin-colistin medium and showed little suppression of the L. pneumophila stock strain. Three of five water samples cultured on CCV agar yielded L. pneumophila, one sample yielded two serogroups of L. pneumophila.
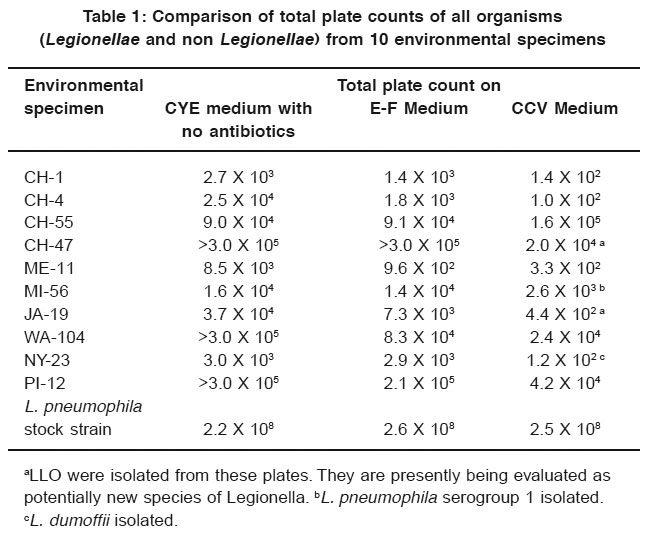 |
Table 1: Comparison of total plate counts of all organisms (Legionellae and non Legionellae) from 10 environmental specimens Click here to view table |
Table 1 lists the isolates of Legionella and LLO and gives the results of the total plate counts of all organisms for the comparison of inhibitory characteristics of CCV, E-F, and CYE media. CCV medium was superior to E-F and CYE media in inhibiting competing flora in the environmental specimens. In addition, Legionella and LLO were isolated from 4 of the 10 samples on CCV medium, whereas no isolations were made from E-F or CYE media.
When CCV plus 80μg of cycloheximide per ml (CCVC medium) was plated with 13 environmental water samples that had already been shown to be positive for Legionella and LLO, 7 were positive on CCVC medium, and only 3 were positive on CYE without antibiotics. However, two of the three isolates on CYEwere recovered from chlorinated water samples relatively free of competing flora.
In the experiments done to develop the acid buffer method, the L. pneumophila stock strain was shown to be resistant to the acid for up to 30 min. The plate count of the stock strain was reduced from 6.3 X 108 colony-forming units per ml at 0 min of acid exposure to 6.1 X 108 at 30 min and to 6.8 X 107 at 60 min. When five environmental samples were exposed to the acid buffer from 30 min by procedure A (see Materials and Methods), L. pneumophila group 1 was recovered from two of the five samples. One of the three samples that did not yield L. pneumophila yielded an LLO isolate. However, two samples tested later yielded isolates of L. pneumophila at 5 min but not at 30 min of exposure to the acid buffer. The total microbial flora for both samples in this instance was reduced by about 2 logs after 5 min of exposure to HCl-KCl buffer, indicating that 5 min is an effective exposure interval for acid buffer treatment.
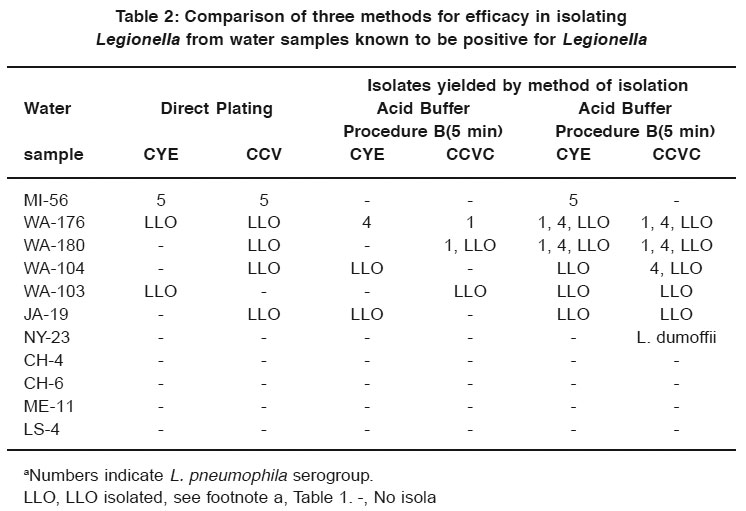 |
Table 2: Comparison of three methods for efficacy in isolating Legionella from water samples known to be positive for Legionella Click here to view table |
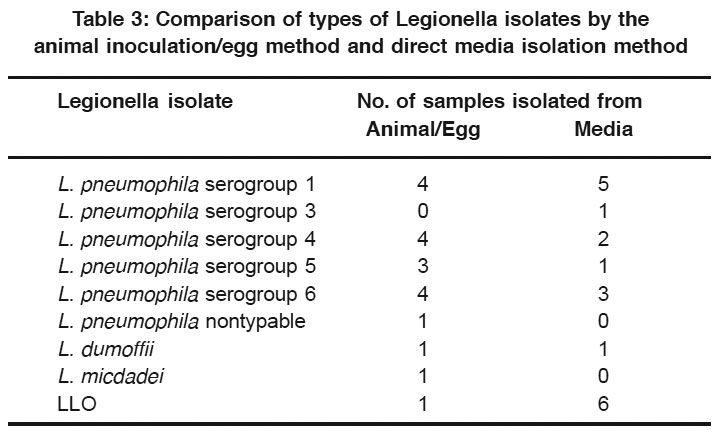 |
Table 3: Comparison of types of Legionella isolates by the animal inoculation/egg method and direct media isolation method Click here to view table |
Table 2 lists the results of the comparison of acid buffer procedures A and B with direct plating onto CYE medium and CCV selective medium. Direct plating yielded isolates of L. pneumophila group 5 from one water sample (MI-56) on both CYE and CCVC. Five of the water samples yielded LLO after direct plating. When procedure B was used, 2 of 11 samples yielded Legionella, and 3 yielded only LLO. When procedure A was used, five water samples yielded Legionella and five yielded LLO.
Analysis of the isolates from 15 environmental samples, tested by both the guinea pig/egg inoculation method and the acid pretreatment/ plating media method discussed above, revealed that multiplespecies and serogroups of Legionella were isolated by both methods (Table 3). The most obvious difference in the groups or species was that LLO was isolatedfrom six samples cultured directlyon plating media, but only one LLO isolate was obtained with the guinea pig/egg inoculation method.
Discussion
Procedure A was superior to both direct plating on CYE and CCVC and to procedure B for isolating Legionella. After the acid buffer treatment, CCVC selective medium used for plating gave slightly better results than CYE medium. However, one L. pneumophila isolate was recovered on CYEthat was not recovered on CCVC medium. This may have been coincidence rather than the result of inhibition of L. pneumophila by the selective medium, but because it occurred we recommend that both CYE and CCVC media be used when samples are pretreated with acid buffer29, 30, 31.
At this time no isolation method for Legionella can completely replace the guinea pig/ egg inoculation method. Since our environmental specimens contained numerous and diverse flora, no single combination of antibiotics tested was effective in suppressing competing flora in all specimens. Even the acid buffer procedure used in combination with CCVC selective medium and CYE medium led to the recovery of Legionella from only 45% of water samples known to contain Legionella. However, it had been as long as a year in some cases between the times Legionella had been isolated from guinea pigs or eggs and the time we did this study. Legionella may not have been still viable in some of the water samples, and the acid pretreatment/ media method may be more effective than our results indicate32, 33, 34, 35.
Direct plating without acid buffer treatment on the selective medium, CCVC agar, led to a recovery rate of up to 36% for Legionella from environmental samples used in our study. It does appear to be more sensitive than animal inoculation for isolation of LLO, possibly indicating that this group of organisms is less pathogenic for guinea pigs (Table 3). These organisms are being considered as possible new Legionella species since they have many characteristics similar to other Legionella.
The utility of CCVC medium for clinical isolation of L. pneumophila and other Legionella species remains to be evaluated. E-F medium, which in our study was not as effective as CCV medium for environmental isolation of Legionella, has been demonstrated to be effective for isolation of L. pneumophila from clinical specimens, although its efficacy for isolation of other species of Legionella is unknown.
Acknowledgements
Authors are thankful to J.C. Bose Institute of Life Science, Department of Biotechnology, Bundelkhand University, Jhansi, 284128 (U.P.), India, for providing financial support and lab facilities for this study.
References
1. Coers J. Monahan C. and Roy C.R., Nat. Cell. Biol., (1999) 1, 451-453.
2. Robinson C.G. and Roy C.R., Cell. Microbiol., (2006) 8, 793-805.
3. Glick T.H. Gregg M.B. Berman B. Mallison G. Rhodes W.W. and Kassanoff I., Am. J. Epidemiol., (1978) 107, 149-160.
4. Joshi A.D. Sturgill-Koszycki S. and Swanson M.S., Cell. Microbiol., (2001) 3, 99-114.
5. Morris G.K. Patton C.M. Feeley J.C. Johnson S.E. Gorman G. Martin W.T. Skaliy P. Mallison G.F. Politi B.D. and Mackel D.C., Ann. Intern. Med., (2001) 90, 664-666.
6. Albert-Weissenberger C. Cazalet C. and Buchrieser C., Cell. Mol. Life Sci., (2007) 64, 432- 448.
7. Segal G. Feldman M. and Zusman T., FEMS Microbiol. Rev., (2005) 29, 65-81.
8. Swanson M.S. and Isberg R.R., Infect. Immun., (1995) 63, 3609 3620.
9. Calvo-Bado L.A. Morgan J.A. Sergeant M. Pettitt T.R. and Whipps J.M., Appl. Environ. Microbiol., (2003) 69, 533-541.
10. Garcia-Fulgueiras A. Navarro C. Fenoll D. Garcia J. Gonzales-Diego P. Jimenez- Bunuelas T. Rodriguez M. Lopez R. Pacheco F. Ruiz J. Segovia M. Balandron B. and Pelaz C. Infect. Dis. (2003) 9, 915-921.
11. Hookey J.V. Saunders N.A. Fry N.K. Birtles R.J. and Harrison T.G., Int. J. Syst. Bacteriol., (1996) 46, 526-531.
12. Sturgill-Koszycki S. and Swanson M.S., J. Exp. Med., (2001) 9, 1261-1272.
13. Hebert G.A. Steigerwalt A.G. and Brenner D.J., Curr. Microbiol., (1980) 3, 255-257.
14. Lewallen K.R. MeKinney R.M. Brenner D.J.Moss C.W. Dail D.H. Thomason B.M. and Bright R.A., Ann. Intern. Med.,(1979) 91, 831-834.
15. Pasculle A.W. Myerowitz R.L. and Rinaldo C.R. Jr., Lancet ii, (1979) 58-61.
16. Den Boer J.W. Yzerman E.P. Schellekens J. Lettinga K.D. Boshuizen H.C. Van Steenbergen J.E. Bosman A. Van den Hof S. Van Vliet H.A. Peeters M.F. Van Ketel R.J. Speelman P. Kool J.L. and Conyn-Van Spaendonck M.A., Emerg. Infect. Dis., (2002) 8, 37- 43.
17. Birtles R.J. Rowbotham T.J. Raoult D. and Harrison T.G., Microbio., (1996) 142, 3525-3530.
18. Nagai H. Kagan J.C. Zhu X. Kahn R.A. and Roy C.R., Sci., (2002) 295, 679-682.
19. Adeleke A. Pruckler J. Benson R. Rowbotham T. Halablab M. and Fields B., Emerg. Infect. Dis., (1996) 2, 225-230.
20. Derré I. and Isberg R.R., Infect. Immun., (2004) 72, 2048-3053.
21. Wadowsky R.M. and Yee R.B., Microbiol., (1981) 42, 768-772.
22. Morrill W.E. Barbaree J.M. Fields B.S.Sanden G.N. and Martin W.T., J. Clin. Microbiol., (1990) 28, 616-618.
23. Seidel K. Bornert W. Baz G. Blankenburg A. and Alexander I., Vom Wasser.,(1986) 67, 39-48.
24. Morris G.K. Steigerwalt A. Feeley J.C. Wong E.S. Martin W.T.. Patton C.M. and Brenner D.J., J. Clin. Microbiol., (1980) 12, 718-721.
25. Amer A.O. and Swanson M.S., Cell. Microbiol., (2005) 7, 765-778.
26. Wang W.L.L. Blaser M.J. Cravens J. and M.A. Johnson., Ann. Intern. Med.(1979) 90, 614-618.
27. Berger K.H. Merriam J.J. and Isberg R.R., Mol. Microbiol., (1994) 14, 809-822.
28. Brand B.C. Sadosky A.B. and Shuman H.A., Mol. Microbiol., (1994) 14, 797-808.
29. Marston B.J. Lipman H.B. and Breiman R.F. Arch. Intern. Med. (1994) 154, 2417-2422.
30. Muder R.R. and Yu V.L., Clin. Infect. Dis., (2002) 35, 990-998.
31. McDade J.E. Shepard C.C. Fraser D.W. Tsai R.R. Redus M.A. and Dowdle W.R., N. Engl. J. Med., (1977) 297, 1197-1203.
32. Verissimo A. Marrao G. da Silva F.G. and da Costa M.S., Environ. Microbiol.,(1991) 57, 2921- 2927.
33. Sheehan K.B. Henson J.M. and Ferris M.J., Appl. Environ. Microbiol., (2005) 71, 507-511.
34. Pascual L. Perez-Luz S. Amo A. Moreno C. Apraiz D. and Catalan V. Can. J. Microbiol., (2001) 47: 341-347.
35. Rowbotham T. J., American Society for Microbio., Washington, D.C. (1993)






